
Meghan Breen
Assistant Professor, Chemistry
- Email: meghan.breen@furman.edu
- Phone: 864.294.2404
- Office: Plyler 236D, Townes Science Center
Dr. Meghan Breen was raised in a military family and grew up living across the US. While growing up she dreamed of becoming a lawyer, but a transformative experience taking two years of high school chemistry led her to pursue a career in chemistry instead. After earning a BS in Chemistry from Eastern Illinois University in 2008, she began pursuing a PhD in Medicinal Chemistry at the University of Michigan. During her graduate work under the guidance of Dr. Matthew Soellner, she developed screening methods to identify ATP-noncompetitive inhibitors of tyrosine kinases. Working as a graduate student instructor was a second transformative experience that led Dr. Breen to pursue a career as a professor at an undergraduate-focused university. After earning her PhD in 2014, she completed a postdoctoral fellowship in chemical biology under the guidance of Dr. Anna Mapp at the University of Michigan Life Sciences Institute. Her postdoctoral research focused on developing chemical tools with applications in chemical and synthetic biology, particularly for the study of transient protein-protein interactions. During her postdoctoral studies, Dr. Breen was named a Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation, and she also taught courses in the Department of Chemistry and Program in Chemical Biolo
Dr. Breen joined the faculty at Furman University in August 2019. Her research program focuses on dissecting the molecular mechanisms that regulate drug resistance in pathogenic fungi and developing new chemical tools for studying transient protein-protein interactions in these organisms. She also is passionate about mentoring and creating opportunities for women and historically excluded groups in STEM. Outside of work, she enjoys traveling, baking, and watching bad TV with her cat.
Honors
- Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation (2016-2019)
- Outstanding Postdoctoral Fellow Award, University of Michigan (2016)
- American Foundation for Pharmaceutical Education Pre-Doctoral Fellowship (2012-2014)
- National Institutes of Health Pharmacological Sciences Training Grant (2009-2011)
- Presidential Scholarship, Eastern Illinois University (2004-2008)
Education
- Postdoctoral Fellowship, Chemical Biology, University of Michigan
- Ph.D. Medicinal Chemistry, University of Michigan
- B.S. Chemistry, Eastern Illinois University
Research
Candida glabrata is the second most common cause of Candida species hospital-acquired bloodstream infections. Both the incidence of C. glabrata infections and the percentage of infections resistant to frontline antifungal drugs have been increasing over the past several decades. The increasing prevalence of drug resistant C. glabrata infections is a growing public health concern, particularly among immunocompromised populations (e.g. transplant recipients, HIV/AIDS patients, premature infants, and cancer patients receiving cytotoxic chemotherapy). Combating anti-fungal resistance will require a better understanding of how drug resistance pathways are regulated and the development of new treatment strategies.
Students in the Breen lab use a variety of biochemical and molecular biology techniques to probe how drug resistance is regulated in C. glabrata. This includes gaining experience culturing and transforming microorganisms, cloning genes and manipulating DNA sequences, and isolating and visualizing proteins. Students also gain experience in scientific communication by presenting their findings at conferences (SERMACS, Protein Society, etc.) and writing a senior thesis.
Regulation of the Pdr1 Transcription Factor by Post-Translational Modifications
One way fungi develop drug resistance is through increased transcription of genes for xenobiotic efflux pump proteins. These proteins expel antifungal drugs from the cell which prevents drugs from accumulating to toxic levels. In C. glabrata, the transcription factor protein Pdr1 initiates the production of xenobiotic efflux pumps implicated in resistance to the azole class of antifungal agents. Knockout of the PDR1 gene results in decreased levels of drug efflux pumps and increased susceptibility to antifungal agents, underscoring the critical role of Pdr1 in regulating drug resistance.
In the Breen lab, we are investigating how post-translational modifications such as phosphorylation regulate Pdr1 and drug resistance in C. glabrata. We have generated mutations in PDR1 gene that prevent post-translational modifications and revert C. glabrata to a drug sensitive phenotype (Figure 1). These Pdr1 variants are being further investigated to characterize how post-translation modifications at specific sites regulate Pdr1 localization and interactions.
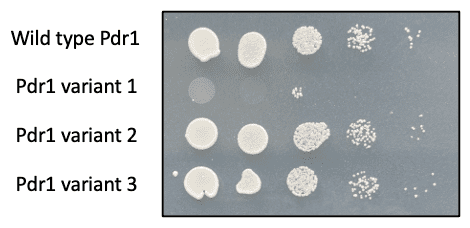
Figure 1. C. glabrata with wild type Pdr1 grows on solid agar medium containing azole drugs. Pdr1 variant 1 reverts C. glabrata to a drug sensitive phenotype, and it no longer survives in the presence of azole drugs.
Genetic Code Expansion in C. glabrata
Normally, proteins are assembled from 20 canonical amino acids, but genetic code expansion techniques incorporate non-canonical amino acids into proteins in a living cell. The non-canonical amino acid can give the protein new chemical properties and enable new types of experiments to probe protein function. Genetic code expansion uses a biorthogonal amino acid tRNA synthetase and tRNA pair to site-specifically incorporate a non-canonical amino acid into a protein at an amber stop codon (UAG). First, the bioorthogonal amino acid tRNA synthetase charges the bioorthogonal tRNA with the non-canonical amino acid, then the bioorthogonal tRNA recognizes the amber codon in mRNA (Figure 2). This converts the amber codon from stop codon to a codon for the non-canonical amino acid. By altering a gene sequence to contain an amber stop codon, a protein can be synthesized with a non-canonical amino acid at a desired location.
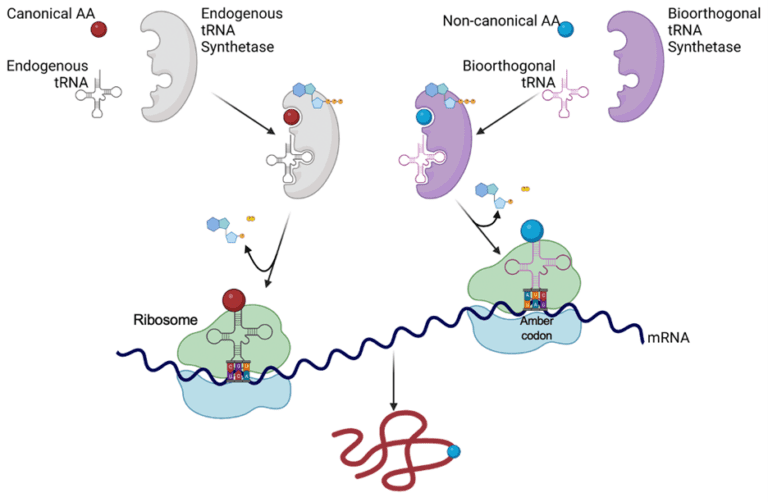
Figure 2. When a bioorthogonal amino acid tRNA synthetase and tRNA are not present, the amber codon acts as a stop codon. If the bioorthogonal amino acid tRNA synthetase, bioorthogonal tRNA, and non-canonical amino acid are present, then the charged bioorthogonal tRNA will recognize the amber codon and the non-canonical amino acid is incorporated into the growing polypeptide chain.
In the Breen lab, we are developing new genetic code expansion systems to produce proteins containing non-canonical amino acids in C. glabrata. While this technique has been used in other species, including the closely related organism Saccharomyces cerevisiae, genetic code expansion has never been utilized in C. glabrata before. The incorporation of non-canonical amino acids will enable us to probe protein-protein interactions regulating drug resistance in their native environment.
Research Funding
- SC-INBRE DRP: Effects of phosphosites on azole resistance in Candida glabrata, 2021-2023.
-
Breen, M. E.; Joy, S. T.; Baruti, O. J.; Beyersdorf, M. S.; Henley, M. J.; DeSalle, S. N.; Ycas, P. D.; Croskey, A.; Cierpicki, T.; Pomerantz, W. C. K.; Mapp, A. K. "Garcinolic Acid Distinguishes Between GACKIX Domains and Modulates Interaction Networks." ChemBioChem, 2023, 24, e202300439.
-
Joiner, C.M.; Breen, M.E.; Mapp, A.K. "Electron‐deficient p‐benzoyl‐L‐phenylalanine derivatives increase covalent chemical capture yields for protein–protein interactions", Protein Science, 2019, 28, 1163-1170.
-
Kersten, K.M.; Breen, M.E.; Mapp, A.K.; Matzger, A.J. Pharmaceutical solvate formation for the incorporation of the antimicrobial agent hydrogen peroxide. Chem. Commun. (Cambridge, U. K.) 2018, 54, 9286-9289.
-
Breen, M.E.; Mapp, A.K. "Modulating the masters: chemical tools to dissect CBP and p300 function", Current Opinion in Chemical Biology, 2018, 45, 195-203.
-
Joiner, C. M.; Breen, M. E.; Clayton, J.; Mapp, A. K. A Bifunctional Amino Acid Enables Both Covalent Chemical Capture and Isolation of in Vivo Protein-Protein Interactions. ChemBioChem 2017, 18, 181-184.
-
Brandvold, K.R.; Santos, S.M.; Breen, M.E.; Lachacz, E.J.; Steffey, M.E.; Soellner, M.B. Exquisitely Specific Bisubstrate Inhibitors of c-Src Kinase. ACS Chem. Biol. 2015, 10, 6, 1387-1391.
-
Breen, M.E.; Soellner, M.B. Small Molecule Substrate Phosphorylation Site Inhibitors of Protein Kinases: Approaches and Challenges. ACS Chem. Biol. 2015, 10, 1, 175-189.
-
Breen, M. E.; Steffey, M. E.; Lachacz, E. J.; Kwarcinski, F. E.; Fox, C. C.; Soellner, M. B. Substrate Activity Screening with Kinases: Discovery of Small‐Molecule Substrate‐Competitive c‐Src Inhibitors. Chem. Int. Ed., 2014, 53 7010-7013.
-
Yestrepsky, B.D.; Yuanxi Xu; Breen, M.E.; Yuanxi Xu; Rajeswaran, W.G.; Ryu, J.G.; Sorenson, R.J.; Tsume, Y.; Wilson, M.W.; Wenpeng Zhang; Duxin Sun; Hongmin Sun; Larsen, S.D. Novel inhibitors of bacterial virulence: Development of 5,6-dihydrobenzo[h]quinazolin-4(3H)-ones for the inhibition of group A streptococcal streptokinase expression. Bioorg. Med. Chem., 2013, 21, 7, 1880-1897.
-
Grove, R.C.; Malehorn, S.H.; Breen, M.E.; Wheeler, K.A. A photoreactive crystalline quasiracemate. Chem. Commun. (Cambridge, U. K.), 2010, 46, 7322-7324.
-
Breen, M.E.; Tameze, S.L.; Dougherty, W.G.; Kassel, W.S.; Wheeler, K.A. Structural Studies of Enantiomers, Racemates, and Quasiracemates. 2-(3-Bromophenoxy)propionic Acid and 2-(3-Methoxyphenoxy)propionic Acid. Cryst. Growth Des. 2008, 8, 10, 3863-3870.